VecScreen (National Center for Biotechnology Information) - screens your DNA sequence for potential vector sequence. Well worth running before doing any other analysis.
Base composition - consider WORDCOUNT (EMBOSS Suite) which gives one the option of choosing the "word size". The latter provides a nice output of mono-, di- and trinucleotide frequencies. Select "create statistics" and "start task" to get to the sequence entry page.
Oligonucleotide frequencies calculator - in addition to calculating the number and frequency of monomeric to octomeric sequences this site also will calculate Zero Order Markov chain for tetranucleotides (ZOM); First Order Markov chain for tetranucleotides (FOM); Second Order Markov chain for tetranucleotides (SOM); and, Z-scores for tetranucleotides.
Compare oligonucleotide frequencies among sequences - This tool will compute distance between input sequences and UPGMA clustering will be display in tabular and dendogram formats.
DNASCANNER v2 - offers an extensive analysis of 158 DNA properties, including mono/di/trinucleotide frequencies, structural, physicochemical, thermodynamics, and mechanical properties of DNA sequences. The tool provides downloadable results and offers interactive plots for easy interpretation and comparison between different features. (Reference: Preeti P et al. 2024. J Comput Biol 31(7): 651-669).
Genomics %G~C Content Calculator (Science Buddies.org) - simple calculator for mol%G+C plus counts the individual bases.
Compositional heterogeneity: GC Content Plot Online. Similar site here.
GraphDNA - DNA Skew Graphing (Viral Bioinformatics Resource Center, University of Victoria, Canada) - this Java applet performs DNA walks, purine, AT and GC skews on small (<1 Mb) genomes. Requires registration and login. Alternative locations for cumulative GC skew are the GC Skewing (Davidson College, U.S.A.). Genomic nucleotide skews are explained here.
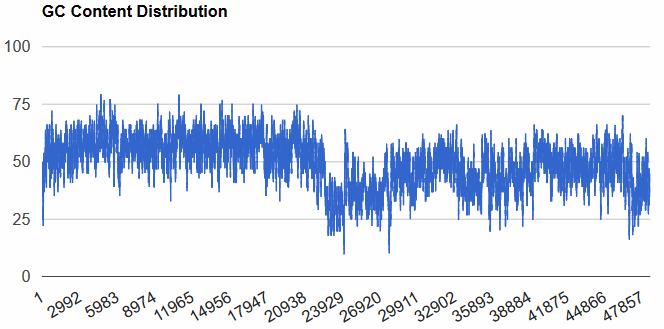
GC Content Calculator (Biologics International Corp, Indianapolis, USA) - DNA GC-content percentage is calculated as Count(G + C)/Count(A + T + G + C) * 100. This program was used to generate the following disgram of Escherichia phage lambda (NC_001416) using a window of 48 bp. One can click on the peaks and valleys and get a read-out of the localized GC-content. Another graphically good site is on the VectorBuilder site.